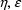 -interaction is now
allowed. This interaction applies to the elements of
-interaction is now
allowed. This interaction applies to the elements of
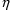 for which conditional
estimates are obtained. Simply include the INTERACTION
option on the $ESTIMATION record. (See the
$ESTIMATION Help
Item.).
for which conditional
estimates are obtained. Simply include the INTERACTION
option on the $ESTIMATION record. (See the
$ESTIMATION Help
Item.).This document provides an introduction to NONMEM Version VI, indicating the new features and basic changes from Version V. Perhaps the most important aspects of Version VI, however, are:
1) all Version V bugs identified as of the date of this document have been fixed,
2) numerous small improvements to Version V have been made, such as messages,
3) the stability and speed of Version V computations have been significantly improved, and depending on the model, size of the data set and estimation method, the user should notice this.
4) Version VI is essentially upwards compatible with Version V. The formatting of the report file, however, is a little different, and there are some minor changes noted below in sections 2.2.5, 4.1.2, 4.1.7, 4.1.15, 4.1.23. Model specification files created by version V cannot be used with version VI and vice versa. There is no single-precision version of NONMEM VI.
5) There is now a single INCLUDE file named SIZES. This file contains all the constants previously contained in NSIZES, PSIZES, TSIZES and LSIZES. SIZES contains a new variable, MAXIDS, the maximum number of individuals in a data set. The default value is 1000.
6) NONMEM Users Guides I-VIII and NONMEM V Supplement are provided as .pdf files on the NONMEM VI CD. With the exception of Guide II, these .pdf files are searchable. At this time, Guides I-VII have not been updated for NONMEM VI. However, Guide VIII, the Help Guide, is fully updated to reflect changes from NONMEM V to NONMEM VI.
7) Updated descriptions of the NONMEM Control Record Formats are provided in an appendix to this document.
NONMEM has three major parts: NONMEM, PREDPP, and NM-TRAN. Strictly speaking, "Version V" refers to the version of NONMEM, where the releases of these parts are V, IV, and III, respectively, and "Version VI" refers to the version of NONMEM, where the releases of these parts are VI, V, and IV, respectively.
Brief descriptions of new features and basic changes from NONMEM Version V that relate to the NONMEM program itself are found in section 2; those that relate to PREDPP are found in section 3; those that relate to NM-TRAN are found in section 4.
Detailed descriptions may be found in the Help items (NONMEM Users Guide VIII). The Help items should also be consulted for some new useful examples. Improvements have been made to many of the older items, in particular NMPRD5 (auto-correlation). Some terminology has been changed in a few help items. What were previously referred to as "scaled, transformed parameters" or STP are now designated as "unconstrained parameters" or UCP.
1. With the Hybrid Estimation method and an
intraindividual mean-variance model (see
NONMEM Users Guide VII ),
the 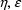 -interaction is now
allowed. This interaction applies to the elements of
-interaction is now
allowed. This interaction applies to the elements of
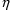 for which conditional
estimates are obtained. Simply include the INTERACTION
option on the $ESTIMATION record. (See the
$ESTIMATION Help
Item.).
for which conditional
estimates are obtained. Simply include the INTERACTION
option on the $ESTIMATION record. (See the
$ESTIMATION Help
Item.).
2. With the LAPLACIAN Estimation method and a
mean-variance model (see
NONMEM Users Guide VII ),
the 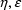 -interaction is now
allowed. In this case the numerical method for obtaining
second partial
-interaction is now
allowed. In this case the numerical method for obtaining
second partial 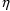 -derivatives
must be used, and therefore too, a "slow"
minimization method will be used. Simply include the
INTERACTION option on the $ESTIMATION record. (See the
Help
Item.)
-derivatives
must be used, and therefore too, a "slow"
minimization method will be used. Simply include the
INTERACTION option on the $ESTIMATION record. (See the
Help
Item.)
3. Background: With the First-Order Estimation
method, if the model is not already linear in
 , it is first approximated by
a first-order Taylor expansion in
, it is first approximated by
a first-order Taylor expansion in
 about
about
 . This approximate model is
then approximated further by a first-order Taylor expansion
in
. This approximate model is
then approximated further by a first-order Taylor expansion
in 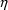 about
about
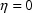 , followed by the
, followed by the
 - interaction term being
dropped. Finally, the Extended Least Squares Estimation
method (1) is used.
- interaction term being
dropped. Finally, the Extended Least Squares Estimation
method (1) is used.
Now the interaction term may be retained,
resulting in the First-Order Estimation method with
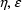 -interaction. Simply include
the INTERACTION option on the $ESTIMATION record. The result
is generally not very different from that obtained when the
interaction term is dropped. Generally, a slightly less
biased estimate of
-interaction. Simply include
the INTERACTION option on the $ESTIMATION record. The result
is generally not very different from that obtained when the
interaction term is dropped. Generally, a slightly less
biased estimate of 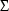 is
obtained, and when there are systematic interindividual
differences in the magnitude of interindividual random
variability itself, a slightly less biased estimate of
is
obtained, and when there are systematic interindividual
differences in the magnitude of interindividual random
variability itself, a slightly less biased estimate of
 is obtained. (See the
$ESTIMATION Help
Item.)
is obtained. (See the
$ESTIMATION Help
Item.)
4. A new population estimation method - called
the Stieltjes Estimation method - is not available in NONMEM
VI 1.0. It may be added to NONMEM VI at a later time. It is
a two-stage method. A population conditional estimation
method should be used to "quickly" obtain fairly
reliable parameter estimates; this comprises the first
stage. Include the METHOD=CONDITIONAL option on the
$ESTIMATION record. The first-stage estimates are used as
initial estimates in the second stage, wherein another
objective function is minimized to obtain a final set of
parameter estimates. The second stage takes much more time
than the first stage, and the resulting parameter estimates
are at least as good as the first-stage estimates, perhaps
better. In the second stage, conditional estimates of
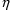 are not used, but the
functions that are minimized to obtain conditional estimates
are still evaluated at many values of
are not used, but the
functions that are minimized to obtain conditional estimates
are still evaluated at many values of
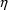 . Generally though,
sufficiently good estimates are obtained by using
conditional estimation methods, or even the First-Order
Estimation method, and one needs to use the Stieltjes
Estimation method only with somewhat pathological data
designs. Simply also include the STIELTJES option (along
with an optional GRID option) on the $ESTIMATION
record.
. Generally though,
sufficiently good estimates are obtained by using
conditional estimation methods, or even the First-Order
Estimation method, and one needs to use the Stieltjes
Estimation method only with somewhat pathological data
designs. Simply also include the STIELTJES option (along
with an optional GRID option) on the $ESTIMATION
record.
5. With minimizations that terminate with the message "MINIMIZATION SUCCESSFUL" in the report file, a warning message will now sometimes also be output:
HOWEVER, PROBLEMS OCCURRED WITH THE MINIMIZATION. REGARD THE RESULTS OF THE ESTIMATION STEP CAREFULLY, AND ACCEPT THEM ONLY AFTER CHECKING THAT THE COVARIANCE STEP PRODUCES REASONABLE OUTPUT.
This may happen when e.g. the final gradient vector is too large.
6. Along with the ETABAR statistic, its estimated standard error is now also output. As with Version V, the corresponding P-value that is output relates to the hypothesis that the true value of ETABAR is 0. With Version VI, this is now a default P-value, and one may obtain an alternative P-value. This is done by implementing a subsequent NONMEM problem which simulates a new data set under the model used with the current problem and obtains parameter estimates for these new data along with an ETABAR statistic, and by requesting that the P-value corresponding to this statistic relate to the hypothesis that the true value of ETABAR for the subsequent problem is the same as that with the original problem. This provides a type of check on the adequacy of the data-analytic model (but the necessity to satisfy this check, like all such checks, is subjective). To get this alternative P-value, generate a model specification file with the original problem, input this file with the subsequent problem, and include the new option ETABARCHECK on the $ESTIMATION record of the subsequent problem. (See the $ESTIMATION and ETABAR Help Items.)
7. With Version V, there are situations where the computation of ETABAR with a mixture model was not sensible. Now multiple ETABAR vectors are output, one for each subpopulation. For each subpopulation, ETABAR is computed over all individuals who have been classified (via a standard Bayesian type computation) to be in the subpopulation. Due to possible misclassification, the default P-value may not be reliable, and it is not output. One may, however, use the alternative P-value described in point 5.
8. A simultaneous analysis of both regular-continuous and odd-type data can be implemented more easily than can be done with Version V. A flag F_FLAG can now be set in a NONMEM-PRED common NMPR17 (via NM-TRAN abbreviated code), which directs NONMEM to interpret the value set in Y (in NM-TRAN abbreviated code) or in F (in a user-supplied PRED or ERROR routine) as being either a "prediction" or a likelihood, on an observation-specific basis. (See the NMPR17 Help Item.)
9. A frequency-prior (2) may be specified for data-analytic purposes, or to simulate parameter values (in a Simulation Step). Two NONMEM utility routines help in this regard. NWPRI allows the THETA vector to be constrained by a "normal shaped prior", and/or the OMEGA matrix to be constrained by an "inverse-Wishart shaped prior", where both priors are specified by the user. TNPRI allows all parameters to be constrained by a prior that arises automatically from a NONMEM analysis of a prior data set. (See the $PRIOR, PRIOR, NWPRI, TNPRI Help Items and the NWPRI, TNPRI examples.)
10. With an intraindividual mean-variance model (see Guide VII), the conditional likelihood for an observation is "conditioned" on values of the population parameters. With Version VI, it may easily be conditioned further; the condition being that the observation is inside or outside a given one- or two-sided interval. This is useful if e.g. blood concentrations below a known quantification limit have been eliminated from the data set. In this case one may use a one-sided interval with lower boundary equal to the quantification limit. It is assumed that the observations of the mean-variance model are normally distributed. With an observation record simply set the boundaries of the interval, YLO and YUP, in the NONMEM-PRED common NMPR12 (via NM-TRAN abbreviated code). (See the NMPR12, and ROCM44 Help Items, and the YLO example.)
11. With an intraindividual mean-variance model
(see Guide VII), the possible values of an observation may
be categorized into one- and/or two-sided intervals, and the
conditional likelihood for an observation may be taken to be
the probability that the observation is in the category into
which it is observed to fall. Categorization may be preceded
by conditioning as described in point 10 above. Interval
boundaries may be random variables. It is assumed that the
observations of the mean-variance model are normally
distributed, so that the probabilities for the categories
are essentially specified by probits. So this feature allows
a probit analysis of data that might otherwise be handled by
a logit analysis. With an observation record, simply set the
lower boundary, CTLO, and upper boundary, CTUP, (and their
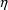 -derivatives) of the
observed interval in the NONMEM-PRED commons NMPR13 and
NMPR14 (via NM-TRAN abbreviated code). (See the
NMPR13,
NMPR14,
ROCM45 Help Items.)
-derivatives) of the
observed interval in the NONMEM-PRED commons NMPR13 and
NMPR14 (via NM-TRAN abbreviated code). (See the
NMPR13,
NMPR14,
ROCM45 Help Items.)
12. The individual contributions to the objective function value are now stored in ROCM50 along with the individual ID values. (See the ROCM50 Help Item.)
13. There is a new option on the $ESTIMATION record. The objective function value and the gradients can be calculated after the individual contributions have been sorted from the lowest to highest absolute values. In some cases this may decrease rounding errors. Simply include the SORT option on the $ESTIMATION record. (See the $ESTIMATION Help Item.) The prototype code for sorting the individual components of the objective function value and the gradients was provided by Dr. Mark Sale, Next Level Solutions, LLC.
1. There are a number of new NONMEM read-only commons, some containing particularly useful variables.
ROCM32: the numbers of the "current" individual record (NIREC) and data record (NDREC) within the individual record
ROCM46: the number of individual records containing at least one observation (NINDR), and the numbers of the first (INDR1) and last (INDR2) such individual records
ROCM48: the total number of data records in the "current" individual record (LIREC)
ROCM49: PRED_, RES_ and WRES_, the PRED, RES and WRES data items, available at problem-finalization (ICALL=3) during any pass through the data set
All these variables are available via NM-TRAN abbreviated code.
(See the ROCM32, ROCM46, ROCM48, ROCM49 Help Items.)
2. Data records are passed sequentially to the PRED routine; heretofore, always from the first data record of an individual record to the last record of the individual record. With Version VI, variables (RPTI, RPTO, RPTON and PRDFL) in NONMEM-PRED common NMPR10 can be set and/or tested (via NM-TRAN abbreviated code), thus allowing a subsequence of data records to be repeatedly passed to PRED multiple times before the next data record following the last record of the subsequence is passed. Subsequences can be nested. This "repetition feature" allows e.g. kinetics to be computed by a convolution integral. The user can exercise control over which pass through a sequence it is, during which the output from PRED with a given data record will be used by NONMEM. This "PRED control" allows e.g. PRED output with a given record to involve computations over subsequent, as well as prior, records. (See the NMPR10 Help Item and the Repetition examples.)
3. The maximum number of subpopulations allowed with a mixture model is now an installation parameter in SIZES. The default value is 4, the maximum number allowed with Version V, and still, a fairly good number to use. With most data sets it is hard to discern more than 4 subpopulations. On the other hand, a recent paper (3) describes a technique for dealing with discrete covariates with missing values, which could greatly increase the required number of mixture subpopulations. (See the SIZES Help Item.)
4. With a mixture model, mixture probabilities can be individual-specific, and so the MIX routine is called repeatedly - with each individual. Useful individual-specific information for setting the probabilities may be made available to MIX by means of the $CONTR record. With Version VI, the data record in NONMEM read-only common ROCM31 serves to provide an additional way for individual-specific information to be made available (and so now serves an additional purpose to that of providing a template data record). With each individual, when MIX is called, the first data record of the individual record may be found in the common. (see Section 4.1.6 and the ROCM31, $CONTR, $MIX Help Items.)
5. With Version V, when a model specification file (MSF) is input, and the Simulation Step is implemented along with other NONMEM steps, the parameter values used in the simulation (the "true values"), as well as the initial parameter values used in conjunction with the other steps, are the final parameter estimates included in the MSF. With Version VI, more flexibility is available with a new option - TRUE - on the $SIMULATION record, which allows the old behavior, but this behavior is not the default behavior. (See the $SIMULATION Help Item.)
6. At problem-finalization, a flag (SKIP_) can now be set in NONMEM-PRED common NMPR15 (via NM-TRAN abbreviated code), which permits a problem (with subproblems), or a superproblem or superproblem iteration to be ended and for NONMEM to continue with the next problem, superproblem or superproblem iteration, respectively. (See the NMPR15 Help Item.)
7. There are a number of new NONMEM-PREDPP commons, some of which are described above. Of particular additional interest:
NMPR16: parameter values produced during the Simulation Step when a frequency prior (see above) is used during that step
8. There is a new step, NONPARAMETRIC. NONMEM obtains either marginal cumulatives or conditional (non-parametric) estimates of etas. (See the $NONPARAMETRIC, $SIMULATION, ROCM18 Help items, and the nonparametric example.)
1. There is a new feature: model event times. These are additional PK parameters MTIME(i) defined in the PK routine or $PK block. A model event time, like an absorption lag time, defines a time to which the system is advanced, but whose value usually cannot be known in advance. When the time is reached, certain indicator variables are set and a call to PK is made. At this call PK, DES, AES and ERROR can use the indicator variables to change some aspect of the system, e.g., a term in a differential equation, or the rate of an infusion. Thus, a model event time can be used e.g. to mark the time at which the gall bladder begins to empty. A model event time could be implemented by using an absorption lag time, and this is what has been done until now, but doing this is somewhat clumsy. There may be up to PCT model event times, where PCT is a new installation parameter defined in SIZES. Its default value is 30. (See the MTIME, $PK, PRDPK1, PRDPK2, Help Items and the MODEL TIME Example)
2. There is a new variable A_0FLG - the
"compartment initialization flag" - in a new
PREDPP read-only common PROCMC. When event records are
passed to routine PK, PREDPP sets A_0FLG to 1 with the first
event record of an individual record or with any reset
record, and to 0 with all other records. When A_0FLG=1,
compartment amounts have been initialized to zero, i.e. the
state vector A (and its 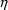 -derivatives) has been set to zero, but PK may reinitialize
the compartment amounts to nonzero values by resetting the
state vector (and its
-derivatives) has been set to zero, but PK may reinitialize
the compartment amounts to nonzero values by resetting the
state vector (and its  -derivatives) to such values (in a new PREDPP-PK common
PRDPK3). (See the $PK,
PROCMC,
PRDPK3,
COMPARTMENT INITIALIZATION BLOCK
Help Items.)
-derivatives) to such values (in a new PREDPP-PK common
PRDPK3). (See the $PK,
PROCMC,
PRDPK3,
COMPARTMENT INITIALIZATION BLOCK
Help Items.)
A_0FLG is a reserved variable in $PK. See section 4.2 below for further discussion as to how to initialize compartment amounts.
3. With the PK routine, the compartment amounts
(and their 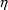 -derivatives) may
be found in PREDPP read-only common PROCM4. These are the
latest computed compartment amounts. That is, they are the
amounts at the previous event time, or if at a later time
(but before the event time for which it may be that PK is
being called) a lagged or additional dose was given, or a
regular infusion was terminated, or an event occurred at a
modeled event time, then they are the amounts at the latest
such time. Now, with Version VI, there is a new PREDPP
read-only common PROCM9 in which the time at which these
compartment amounts are in fact computed may be found. (See
the PROCM4,
PROCM9 Help Items.)
-derivatives) may
be found in PREDPP read-only common PROCM4. These are the
latest computed compartment amounts. That is, they are the
amounts at the previous event time, or if at a later time
(but before the event time for which it may be that PK is
being called) a lagged or additional dose was given, or a
regular infusion was terminated, or an event occurred at a
modeled event time, then they are the amounts at the latest
such time. Now, with Version VI, there is a new PREDPP
read-only common PROCM9 in which the time at which these
compartment amounts are in fact computed may be found. (See
the PROCM4,
PROCM9 Help Items.)
4. When the value of the compartment data item (CMT) is n+1, where n is the total number of compartments in the kinetic system, the data item designates the (special additional) output compartment. With Version VI, this can also be done with the value 100, which is useful when n changes from run to run. (See the CMT Help Item.)
1. The IGNORE option on the $DATA record has been expanded so that any data record will be dropped when the values of one or more data items meet certain user-specified criteria, and a new ACCEPT option has been introduced so that any data records will be dropped unless the values of one or more data items meet certain user-specified criteria. (See the $DATA Help Item.)
2. With Version V, the option TRANSLATE=(TIME/24) (or TRANSLATE=(II/24)) on the $DATA record results in the TIME (or II) data items in units of hours being converted to data items in units of days, with two figures to the right of the decimal point being retained. With version VI, this option results in three figures to the right of the decimal point being retained, as does the option TRANSLATE=(TIME/24.000) (or TRANSLATE=(II/24.000)). The option TRANSLATE=(TIME/24.00) (or TRANSLATE=(II/24.00)) results in two figures to the right of the decimal point being retained. (See the $DATA Help Item.)
3. With Version V, the DO WHILE construction is allowed in an initialization/finalization block, and when this is used in conjunction with direct calls to the NONMEM utility routine PASS, passes through the data set may be implemented during problem initialization/finalization. Now a new DO WHILE (DATA) construction is allowed, which automatically and transparently manages the calls to PASS. (See the ABBREVIATED CODE and PASS Help Items.)
4. With Version V, a random variable defined in an abbreviated code for a given routine must be completely defined with each data record that is passed to the routine. NM-TRAN now supports recursive definition of random variables in $PRED, $PK and $ERROR blocks. Examples of recursively defined random variables are given in the following fragments of code.
Example 1 - random variable retains value from
previous data record.
This example illustrates how one can use abbreviated code
when WT is included only on some (e.g. the first) data
records of an individual record. (Of course, this can lead
to problems with developing plots involving WT.) Here, K is
defined in terms of ETA(1), and so it is a random
variable.
IF (WT.GT.0) THEN
K=THETA(1)*WT*EXP(ETA(1))
ELSE
K=K
ENDIF
Example 2 - bolus dose at time 0
This example illustrates how one can use abbreviated code to
implement recursive kinetics in $PRED assuming that TIME is
the elapsed time since the previous data record.
K=THETA(1)*EXP(ETA(1))
IF (TIME.EQ.0) OLDA=AMT
A=OLDA*EXP(-K*TIME)
OLDA=A
Example 3 - multiple bolus doses
This example further illustrates how one can use abbreviated
code to implement recursive kinetics in $PRED assuming that
TIME is the elapsed time since the previous data
record.
K=THETA(1)*EXP(ETA(1))
IF (TIME.EQ.0) THEN
A=AMT
ELSE
A=A*EXP(-K*TIME)+AMT
ENDIF
5. The variables mentioned in sections 2 and 3: F_FLAG, YLO, YUP, CTLO, CTUP, NIREC, NDREC, NINDR, INDR1, INDR2, LREC, PRED_, RES_, WRES_, RPTI, RPTO, RPTON, PRDFL, and SKIP_ are all available with abbreviated code, as described in those sections.
6. Other variables that now may be used (as right-hand quantities) in abbreviated code are:
NEWL2 (from common ROCM17)
TEMPLT(n) (from common ROCM31)
The TEMPLT(n) variables refer to the items of the template data record. They may be used in data average blocks (ICALL=6) and $MIX blocks. The items of the template record may be referred to by position or by label, e.g., TEMPLT(1) or TEMPLT(ID). (See ROCM17 and ROCM31 Help Items.)
7. With Version VI, the variables S1SC and S2SC of NONMEM read-only common ROCM14 are no longer recognized in initialization/finalization blocks; rather, the variables S1NUM and S2NUM are used. (See ROCM14 and $SUPER Help Items.)
8. User-written functions FUNCA, FUNCB and FUNCC (called abbreviated functions) may now be used as right-hand quantities in abbreviated codes. Such a function may depend on arguments which may be random variables, in which case the function too becomes a random variable. Elements of vectors (VECTRA(n), VECTRB(n), VECTRC(n)) may be defined as left-side quantities in abbreviated code. A vector may be used as an argument to an abbreviated function. The code for the function itself must be written by the user in FORTRAN. (See the ABBREVIATED FUNCTION Help Item.)
9. The functions TAN, ASIN (arcsin), ACOS (arccosine), ATAN (arctangent) may now be used in abbreviated code. The INT function may also now be used. When this function is used outside a simulation block, INT of a random variable is regarded as a non-random variable.
10. The NONMEM utility routine RANDOM may now be used in initialization/finalization blocks.
11. The value of the RECORDS option of the $DATA record may now be a label (appearing in the $INPT record) of a data item type. In particular, it may be ID (alternatively, IR, INDREC, or any initial substring of length 3 or more of INDIVIDUALRECORD). With this value NM-TRAN understands the data records for the problem to start with the first data record of the NM-TRAN data set (from the place where the NM-TRAN data set is positioned before data records are read; see the NOREWIND option) and to include all data records in the series of contiguous data records that follow and having the same value of the data item as does the first record. It counts the number of these data records (minus comment or dropped records) and puts this number in the NONMEM control file, so that these data records (or NM-TRAN-modified versions of them) become the data records used in the problem. (See the $DATA Help Item.)
There are two obvious applications. First, during a checkout phase, one might want to see only the predictions for the first individual record, and therefore, use a data set comprised only of the data records therein. When RECORDS=ID is used, one need not count these data records. Second, one might want to analyze each subject’s data independently of the others. One way this can be done is by creating a single control stream with as many problems as there are individual records in the data, and including one each of the following series of $DATA records in the series of problem specifications:
$DATA filename RECS=n1 $DATA filename RECS=n2 NOREWIND $DATA filename RECS=n3 NOREWIND etc.
where "nk" is the number of records in the kth individual record. It would be necessary to count the exact number of records. Now, these records can simply be coded
$DATA filename RECS=ID $DATA filename RECS=ID NOREWIND $DATA filename RECS=ID NOREWIND etc.
(See $DATA Help Item.)
12. With the INCLUDE record, the filename may now be followed by an integer n, whose default value is 1. Then NM-TRAN reads n copies of the named file.
If in the example above, where the problem specification for all subjects after the first are completely identical, a compact way of writing the control stream is possible. The control stream would contain the problem specification for the first individual, including the $SUBROUTINES record, abbreviated code, and the $DATA record for the first subject, and would end with (if there are e.g. 12 subjects in all)
INCLUDE ctlfile2 11
The file ctlfile2 would contain one problem specification for one subject, with no $SUBROUTINES record or abbreviated code, but would include
$DATA filename RECS=ID NOREWIND
(See INCLUDE Help Item.)
13. The INTERACTION option may now be used on the $ESTIMATION record along with METHOD=HYBRID, or METHOD=CONDITIONAL, LAPLACIAN, or METHOD=ZERO (see section 2.1). The ETABARCHECK option may be used on the $ESTIMATION record (see section 2.1). (See $ESTIMATION Help Item.)
14. The TRUE option may be used on the $SIMULATION record (see section 2.2 and the $SIMULATION Help Item).
15. With Version V, when the data are single-subject and the L2 data item is defined, NM-TRAN creates new data items and labels them .ID.. Now the ID data item is taken to be the L2 data item, and new .ID. items are not created. The ID item may be defined, but then it is not in turn identified to NONMEM as such. The labels L2 and ID (if defined) may be used in abbreviated code, and then they refer to the corresponding items of the data record, as specified in the $INPUT record. (See the L2 Help Item.)
However, if, as previously, the user uses the label L1 instead of the label ID, or uses the label L1 as a synonym with the ID label, then NM-TRAN does not change the designation: For NONMEM, the items labeled L1 are taken to be the ID data items. (See the ID and L2 Help Items.
16. OMEGA(i,j) and SIGMA(i,j), where the
subscripts i and j are integer constants, can now be used as
right-hand quantities in $PRED, $PK, $ERROR and $INFN
abbreviated codes For example, when PREDPP is used with a
slope-intercept residual error model, the computation of
individual weighted residuals (with F evaluated at
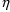 =0) could be done as
follows:
=0) could be done as
follows:
$ERROR Y=F+EPS(1)+F*EPS(2) IF (COMACT.EQ.1) THEN STD=SQRT(SIGMA(1)+F**2*SIGMA(2)) IWRES=(DV-F)/STD ENDIF
When the two subscripts i and j are the same, only one subscript is needed. The OMEGA and SIGMA elements may be listed in WRITE and PRINT statements in initialization and finalization blocks. (See the ROCM22, INITIALIZATION-FINALIZATION BLOCK, and WRITE/PRINT Help Items.)
17. SETHET(i,j), SEOMEG(i,j) and SESIGM(i,j), where the subscripts are integer constants, can now be listed in WRITE or PRINT statements in finalization blocks. These are elements of the estimated standard error arrays for the final estimates of THETA, OMEGA and SIGMA. (See ROCM7 Help Item.)
18. Each of the array names THETA, OMEGA, SIGMA, SETHET, SEOMEG, SESIGM, and ETA - without subscripts, may be listed in a WRITE or PRINT statement, and then the entire array is written. With a WRITE statement, the format should either be omitted or be "*"; NM-TRAN creates a suitable format. E.g., either of
WRITE (99) THETA,SETHET WRITE (99,*) THETA,SETHET
may be used. In such a statement, only arrays may be listed, and only if these arrays share the same dimensions. The array names THETA and ETA may be listed within any block of abbreviated code in which the elements of these arrays may appear on the right (e.g. $PRED and $INFN). The array names OMEGA and SIGMA may be listed within any initialization/finalization block. The array names SETHET, SEOMEG and SESIGM may be listed within any finalization block.
With arrays OMEGA, SEOMEG, SIGMA and SESIGM, the option (BLOCK) or (DIAG) must be given to indicate if the full array or just the diagonal elements are to be written. If the (BLOCK) option is used, the array is displayed in full symmetric form. E.g.,
IF (ICALL.EQ.3) WRITE (99) OMEGA(BLOCK)
may be used.
(See the ROCM7, ROCM22, INITIALIZATION-FINALIZATION BLOCK, and WRITE/PRINT Help Items.)
19. Character constants may be included in WRITE and PRINT statements in abbreviated code. E.g.
PRINT *, ’CLEARANCE EQUALS: ’, CLE
(See the WRITE/PRINT "Help Item
20. The statements OPEN(u), CLOSE(u), REWIND(u), where u is a I/O logical unit number, are now useable in abbreviated codes. The unit number must be an integer constant between 40 and 99. A file name may be included with the OPEN statement, i.e. OPEN(u,FILE=filename). With OPEN and CLOSE, the NONMEM utility routine FILES is automatically called. (See the Abbreviated Code and FILES Help Items.)
21. Copying blocks of abbreviated code begin with tests of the reserved variable COMACT, as follows:
IF (COMACT.EQ....) THEN .... ENDIF
Unlike with Version V,
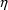 -derivatives are no longer
computed in a copying block. COM(i) variables that are used
on the left in a copying block are referred to as
"explicit" save variables. They are automatically
stored in the SAVE region (a subregion of the reserved
region of common NMPRD4). When COM(i) variables are defined
in a copying block, the reserved region of common NMPRD4
must be explicitly set up with the use of the COMRES option
on the $ABBREV record, but the COMSAV option need not be
used.
-derivatives are no longer
computed in a copying block. COM(i) variables that are used
on the left in a copying block are referred to as
"explicit" save variables. They are automatically
stored in the SAVE region (a subregion of the reserved
region of common NMPRD4). When COM(i) variables are defined
in a copying block, the reserved region of common NMPRD4
must be explicitly set up with the use of the COMRES option
on the $ABBREV record, but the COMSAV option need not be
used.
Variables other than COM(i) variables may be used on the left in a copying block, and they are referred to as "implicit" save variables. These may not be defined along with COM(i) variables. Implicit save variables too are automatically stored in the SAVE region, and in this case, a reserved region of common NMPRD4 is automatically created; neither the COMRES nor COMSAV options can be used on the $ABBREV record. Use of implicit save variables can obviate the need for a $ABBREV record. (See the COM COMACT COMSAV COMRES Help Item.)
22. A new NM-TRAN record concerns the computation of raw data averages. Heretofore, a data item occuring in the data record after the DV data item was not used in template matching. Now, by default it is. If a data item is to be excluded from template matching, the label or synonym for this item should be included in the new record - $OMIT. E.g.
$OMIT TIME
As always, data items required by NONMEM are automatically excluded from template matching. The label of such an item may be included in the $OMIT record, but this is unnecessary. (See the $OMIT Help Item.)
23. With Version V, the OTHER option on the $SUBROUTINES record could be used at most three times. Now it can be used at most forty times.
1. The variable A_0FLG is a new reserved variable in $PK abbreviated code, which may be tested as a right-hand quantity (see section 3). When A_0FLG=1, compartment amounts may be initialized:
IF (A_0FLG.EQ.1) THEN compartment initialization block ENDIF
A_0(i) also is now a reserved variable which may be used as a left-hand quantity in a compartment initialization block. Specifically, A_0(i) is the compartment initialization value of A(i), i.e., the value to which the ith compartment amount is to be set when A_0FLG=1. A_INITIAL(i) is a synonym for A_0(i). The above is referred to as an explicit use of A_0FLG.
There may be implicit uses. (See the COMPARTMENT INITIALIZATION BLOCK Help Item.)
2. Variables A(n) may now be used as right-hand quantities in $PK abbreviated code, where they denote the latest computed compartment amounts. That is, the A(n) are the amounts at the previous event time, or if at a later time (but before the event time for which it may be that PK is being called) a lagged or additional dose was given, or a regular infusion was terminated, or an event occurred at a modeled event time, then the A(n) are the amounts at the latest such time. The time at which the A(n) are in fact computed is found in the variable TSTATE (from PREDPP read-only common PROCM9), also a right-hand quantity in $PK.
If ETA variables and A(n) variables are used in $PK abbreviated code, then any $OMEGA records referring to these ETA variables should precede the $PK record, or if an $MSFI record is used, this record should precede the $PK record and include the option NPOP=m, where m is the number of population ETA variables. (See the $PK and PROCM9 Help Items.)
3. A new NM-TRAN record, $INFN, allows abbreviated code to substitute for user-written FORTRAN code. NM-TRAN generates the FORTRAN INFN routine (there is also an NM-TRAN Library INFN routine). In an $INFN block, ICALL may be tested for initialization/finalization values 0, 1 and 3:
IF (ICALL.EQ.0) THEN run initialization block ENDIF IF (ICALL.EQ.1) THEN problem initialization block ENDIF IF (ICALL.EQ.3) THEN problem finalization block ENDIF
ICALL may now also be tested for these values in $PK and $ERROR blocks, in which case the run initialization, problem initialization, or problem finalization block is moved to an INFN routine. (See $INFN and INITIALIZATION-FINALIZATION BLOCK Help Items.)
1. Beal, SL. Populations pharmacokinetic data and parameter estimation based on their first two statistical moments. Drug Metabolism Reviews 1984; 15: 173-193.
2. Gisleskog, PO, Karlsson, MO, Beal SL. Use of prior information to stabilize a population data analysis. J. Pharmacokin. Pharmacodyn. 2002; 29: 473-505.
3. Beal SL. Conditioning on certain random events associated with statistical variability in PK/PD. J. Pharmacokin. Pharmacodyn. 2005; 32: 213-243.